

Immune responses are orchestrated by controlling the initiation, magnitude, and duration of various signaling pathways. Adaptor proteins act as positive or negative regulators by targeting critical molecules of signaling cascades. Signal-transducing adaptor protein-2 (STAP-2) contains typical features of adaptor proteins, like a pleckstrin homology (PH) domain in the N-terminal region and a Src homology 2 (SH2) domain in the central region. STAP-2 binds to a variety of signaling or transcriptional molecules to control multiple steps of inflammatory/immune responses. STAP-2 enhances T-cell receptor (TCR)-mediated signaling via the association with TCR-proximal CD3ζ immunoreceptor tyrosine-based activation motifs (ITAMs) and lymphocyte-specific protein tyrosine kinase (Lck). STAP-2 decreases adherence of T-cells to fibronectin (FN) through an association with focal adhesion kinase (Fak) and Casitas B-lineage Lymphoma (c-Cbl), and increases chemotaxis of T-cells toward stromal cell-derived factor-1α (SDF-1α) through interactions with Vav1 and Ras-related C3 botulinum toxin substrate 1 (Rac1). STAP-2 positively regulates activation-induced cell deathrough the association with Fas and caspase-8. This review describes the current knowledge of the roles of STAP-2 in T-cell-dependent immune responses and the possible clinical utility of STAP-2-targeting therapies.
Immune responses are orchestrated by controlling the initiation, magnitude, and duration of various signaling pathways. Adaptor proteins act as positive or negative regulators by targeting critical molecules of signaling cascades. Signal-transducing adaptor protein-2 (STAP-2) contains typical features of adaptor proteins, like a pleckstrin homology (PH) domain in the N-terminal region and a Src homology 2 (SH2) domain in the central region. STAP-2 binds to a variety of signaling or transcriptional molecules to control multiple steps of inflammatory/immune responses. STAP-2 enhances T-cell receptor (TCR)-mediated signaling via the association with TCR-proximal CD3ζ immunoreceptor tyrosine-based activation motifs (ITAMs) and lymphocyte-specific protein tyrosine kinase (Lck). STAP-2 decreases adherence of T-cells to fibronectin (FN) through an association with focal adhesion kinase (Fak) and Casitas B-lineage Lymphoma (c-Cbl), and increases chemotaxis of T-cells toward stromal cell-derived factor-1α (SDF-1α) through interactions with Vav1 and Ras-related C3 botulinum toxin substrate 1 (Rac1). STAP-2 positively regulates activation-induced cell deathrough the association with Fas and caspase-8. This review describes the current knowledge of the roles of STAP-2 in T-cell-dependent immune responses and the possible clinical utility of STAP-2-targeting therapies.
DOI: https://doi.org/10.37349/ei.2022.00082
This article belongs to the special issue The Role of Adaptor Proteins in Lymphoid Cell Signaling

Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), a highly pathogenic β-coronavirus, is the etiologic agent of coronavirus disease 2019 (COVID-19), which gave rise to a difficult to control pandemic, especially in Brazil. Approximately 4,000 mutations have been identified in SARS-CoV-2, with the majority being redundant without having any biological effect on the virus. The aim of the present study was to objectively understand how new SARS-CoV-2 variants can affect vaccine response, in addition to highlighting the current situation in Brazil in the face of the pandemic and considering epidemiological and immunological aspects of COVID-19. The main protective correlate investigated in most vaccines is the neutralizing antibody titer induced by immunizing agents, observed in the pre-clinical phase in animals, whose action is to block the binding of the spike protein to the angiotensin-converting enzyme 2 (ACE2) receptor, preventing infection. Up to the second half of 2021, the variants that are of greatest concern worldwide and require molecular surveillance are Alpha variant (or B.1.1.7 lineage), Beta (or B.1.351 lineage), Gamma (or P1 lineage) and Delta (or B.1.617.2 lineage). Brazil finds itself in a highly unfavorable scenario, with the circulation of variants of concern, mainly Gamma and Delta, with high fatality rates for COVID-19 and low vaccination rate. Given the still latent situation of the COVID-19 pandemic in Brazil, the lack of global planning for action strategies for non-pharmacological prevention measures, there is an imminent risk of the emergence of new variants due to the finding of susceptible hosts and the high proliferative rate of SARS-CoV-2. It is urgent to increase the genotyping of positive samples isolated from infected individuals, the speed of vaccination of the entire population and the unification of non-pharmacological preventive measures throughout the country.
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), a highly pathogenic β-coronavirus, is the etiologic agent of coronavirus disease 2019 (COVID-19), which gave rise to a difficult to control pandemic, especially in Brazil. Approximately 4,000 mutations have been identified in SARS-CoV-2, with the majority being redundant without having any biological effect on the virus. The aim of the present study was to objectively understand how new SARS-CoV-2 variants can affect vaccine response, in addition to highlighting the current situation in Brazil in the face of the pandemic and considering epidemiological and immunological aspects of COVID-19. The main protective correlate investigated in most vaccines is the neutralizing antibody titer induced by immunizing agents, observed in the pre-clinical phase in animals, whose action is to block the binding of the spike protein to the angiotensin-converting enzyme 2 (ACE2) receptor, preventing infection. Up to the second half of 2021, the variants that are of greatest concern worldwide and require molecular surveillance are Alpha variant (or B.1.1.7 lineage), Beta (or B.1.351 lineage), Gamma (or P1 lineage) and Delta (or B.1.617.2 lineage). Brazil finds itself in a highly unfavorable scenario, with the circulation of variants of concern, mainly Gamma and Delta, with high fatality rates for COVID-19 and low vaccination rate. Given the still latent situation of the COVID-19 pandemic in Brazil, the lack of global planning for action strategies for non-pharmacological prevention measures, there is an imminent risk of the emergence of new variants due to the finding of susceptible hosts and the high proliferative rate of SARS-CoV-2. It is urgent to increase the genotyping of positive samples isolated from infected individuals, the speed of vaccination of the entire population and the unification of non-pharmacological preventive measures throughout the country.
DOI: https://doi.org/10.37349/ei.2021.00029
This article belongs to the special issue Vaccine-induced Immune Responses Against SARS-CoV-2 Infections

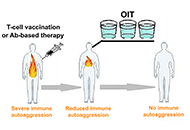
Immunotherapeutic treatment of autoimmune diseases should aim to inactivate autoaggressive memory T-cells and restore immune tolerance. It is envisaged that three approaches could be used to achieve this goal: stimulation of anti-idiotypic immune responses by vaccination with pathogenic T-cells; administration of suboptimal doses of antibodies (Abs) against two or more surface T-cell markers to provide selective Ab-mediated destruction of activated pathogenic memory T-cells; and induction of oral immune tolerance. The proposal entails the use of T-cell vaccination (TCV) or Ab-based therapy as an initial approach to reduce autoantigenic T-cell sensitization. Subsequently, the implementation of oral immunotherapy (OIT) is recommended to reinstate a consistent immune tolerance.
Immunotherapeutic treatment of autoimmune diseases should aim to inactivate autoaggressive memory T-cells and restore immune tolerance. It is envisaged that three approaches could be used to achieve this goal: stimulation of anti-idiotypic immune responses by vaccination with pathogenic T-cells; administration of suboptimal doses of antibodies (Abs) against two or more surface T-cell markers to provide selective Ab-mediated destruction of activated pathogenic memory T-cells; and induction of oral immune tolerance. The proposal entails the use of T-cell vaccination (TCV) or Ab-based therapy as an initial approach to reduce autoantigenic T-cell sensitization. Subsequently, the implementation of oral immunotherapy (OIT) is recommended to reinstate a consistent immune tolerance.
DOI: https://doi.org/10.37349/ei.2023.00117

Aim:
Atopic dermatitis (AD) is a pruritic, chronic inflammatory skin disease. Thymic stromal lymphopoietin (TSLP) is highly expressed in the epidermis of patients with AD and induces T helper 2 (Th2) immune responses and itching. Although the mechanism underlying the stimulus-induced TSLP production in normal keratinocytes has been intensively studied, whether the production capability of TSLP is naturally enhanced in epidermal cells in AD conditions remains unclear. Previous studies demonstrated that a deficiency of polyunsaturated fatty acid (PUFA) causes AD-like pruritic skin inflammation in special diet-fed hairless mice. The aim of the study was to examine the TSLP production capability of epidermal cells isolated from diet-induced AD mouse model and its mechanism.
Methods:
Epidermal cells were isolated from normal and AD mice and incubated under unstimulated culture conditions to assess spontaneous TSLP production. Messenger ribonucleic acid (mRNA) and protein levels of TSLP were determined by real-time polymerase chain reaction (PCR) and enzyme-linked immunosorbent assay (ELISA), respectively.
Results:
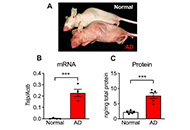
TSLP level was markedly increased in the skin of AD mice. When epidermal cells were isolated from AD mice and cultured without stimulation, Tslp gene expression was upregulated, and a large amount of TSLP protein was extracellularly released. Such TSLP overproduction was not observed in the epidermal cells of normal mice. TSLP overproduction in AD epidermal cells was almost completely inhibited by extracellular calcium chelation, interference with plasma membrane interaction of stromal interaction molecule 1 (STIM1), blockade of the calcium release-activated calcium (CRAC) channels Orai1 and Orai2, or treatment with a PUFA γ-linolenic acid (GLA).
Conclusions:
Epidermal cells isolated from AD mice can spontaneously produce TSLP through STIM/Orai-mediated calcium entry, and GLA may negatively regulate this TSLP production.
Aim:
Atopic dermatitis (AD) is a pruritic, chronic inflammatory skin disease. Thymic stromal lymphopoietin (TSLP) is highly expressed in the epidermis of patients with AD and induces T helper 2 (Th2) immune responses and itching. Although the mechanism underlying the stimulus-induced TSLP production in normal keratinocytes has been intensively studied, whether the production capability of TSLP is naturally enhanced in epidermal cells in AD conditions remains unclear. Previous studies demonstrated that a deficiency of polyunsaturated fatty acid (PUFA) causes AD-like pruritic skin inflammation in special diet-fed hairless mice. The aim of the study was to examine the TSLP production capability of epidermal cells isolated from diet-induced AD mouse model and its mechanism.
Methods:
Epidermal cells were isolated from normal and AD mice and incubated under unstimulated culture conditions to assess spontaneous TSLP production. Messenger ribonucleic acid (mRNA) and protein levels of TSLP were determined by real-time polymerase chain reaction (PCR) and enzyme-linked immunosorbent assay (ELISA), respectively.
Results:
TSLP level was markedly increased in the skin of AD mice. When epidermal cells were isolated from AD mice and cultured without stimulation, Tslp gene expression was upregulated, and a large amount of TSLP protein was extracellularly released. Such TSLP overproduction was not observed in the epidermal cells of normal mice. TSLP overproduction in AD epidermal cells was almost completely inhibited by extracellular calcium chelation, interference with plasma membrane interaction of stromal interaction molecule 1 (STIM1), blockade of the calcium release-activated calcium (CRAC) channels Orai1 and Orai2, or treatment with a PUFA γ-linolenic acid (GLA).
Conclusions:
Epidermal cells isolated from AD mice can spontaneously produce TSLP through STIM/Orai-mediated calcium entry, and GLA may negatively regulate this TSLP production.
DOI: https://doi.org/10.37349/ei.2023.00096
This article belongs to the special issue Cytokines and Skin Diseases

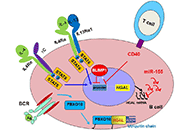
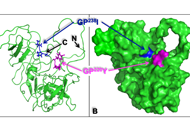
Human germinal center (GC)-associated lymphoma (HGAL) is a multi-domain adaptor protein expressed in GC B lymphocytes, T follicular helper (Tfh) cells and lymphomas derived from these cells. HGAL expression is an independent predictor of longer survival of diffuse large B-cell lymphoma (DLBCL) and classical Hodgkin’s lymphoma (HL) patients. HGAL regulates B cell receptor (BCR) signaling and immunological synapse formation by binding to either the downstream effectors [e.g., spleen tyrosine kinase (Syk)] or other signaling regulators [e.g., growth factor receptor-bound protein 2 (Grb2)]. HGAL regulates the cytoskeleton that reshapes B cell morphology during BCR signaling and cell motility by at least two molecular mechanisms: enhanced Ras homolog gene family member A (RhoA) signaling and inhibition of myosin-actin translocation. These effects on the cytoskeleton decrease lymphoma dissemination in animal models and contribute to decreased lymphoma dissemination in patients. The latter may contribute to the association of HGAL protein expression with longer survival of patients with DLBCL and HL tumors. The ability to regulate multiple and distinct functions simultaneously in B cells implies that the HGAL protein level is tightly regulated. It was demonstrated that HGAL can be regulated by PR/SET domain 1 (PRDM1)/B lymphocyte-induced maturation protein-1 (BLIMP1) and interleukin-4 (IL-4) at the transcription level, by microRNA-155 (miR-155) at the post-transcriptional level, and by F-box protein 10 (FBXO10) at the post-translational level. Constitutive enforced expression of HGAL at physiological levels leads to lymphoid hyperplasia and DLBCL in mice. Future studies need to focus on identifying HGAL interactome, dissecting its interaction network, and understanding HGAL spatiotemporal signaling in live cells in physiological conditions. Further, the recent demonstration of HGAL expression in Tfh cells requires the determination of its function in these cells. These studies will contribute to new insights into the biology of these cellular subsets and how immune dysregulation contributes to lymphomagenesis.
Human germinal center (GC)-associated lymphoma (HGAL) is a multi-domain adaptor protein expressed in GC B lymphocytes, T follicular helper (Tfh) cells and lymphomas derived from these cells. HGAL expression is an independent predictor of longer survival of diffuse large B-cell lymphoma (DLBCL) and classical Hodgkin’s lymphoma (HL) patients. HGAL regulates B cell receptor (BCR) signaling and immunological synapse formation by binding to either the downstream effectors [e.g., spleen tyrosine kinase (Syk)] or other signaling regulators [e.g., growth factor receptor-bound protein 2 (Grb2)]. HGAL regulates the cytoskeleton that reshapes B cell morphology during BCR signaling and cell motility by at least two molecular mechanisms: enhanced Ras homolog gene family member A (RhoA) signaling and inhibition of myosin-actin translocation. These effects on the cytoskeleton decrease lymphoma dissemination in animal models and contribute to decreased lymphoma dissemination in patients. The latter may contribute to the association of HGAL protein expression with longer survival of patients with DLBCL and HL tumors. The ability to regulate multiple and distinct functions simultaneously in B cells implies that the HGAL protein level is tightly regulated. It was demonstrated that HGAL can be regulated by PR/SET domain 1 (PRDM1)/B lymphocyte-induced maturation protein-1 (BLIMP1) and interleukin-4 (IL-4) at the transcription level, by microRNA-155 (miR-155) at the post-transcriptional level, and by F-box protein 10 (FBXO10) at the post-translational level. Constitutive enforced expression of HGAL at physiological levels leads to lymphoid hyperplasia and DLBCL in mice. Future studies need to focus on identifying HGAL interactome, dissecting its interaction network, and understanding HGAL spatiotemporal signaling in live cells in physiological conditions. Further, the recent demonstration of HGAL expression in Tfh cells requires the determination of its function in these cells. These studies will contribute to new insights into the biology of these cellular subsets and how immune dysregulation contributes to lymphomagenesis.
DOI: https://doi.org/10.37349/ei.2023.00097
This article belongs to the special issue The Role of Adaptor Proteins in Lymphoid Cell Signaling

Periodontal tissue destruction can cause complaints for sufferers. Inflammatory conditions of the gingiva, bleeding gums, and even tooth loss are clinical features of the destruction of the periodontal tissues. Periodontitis is an inflammatory disease involving the periodontal tissues. The prevalence of periodontium destruction increases with aging. Changes in innate and adaptive immunity that occur in the elderly also play a role in the severity of periodontitis. “Inflammaging” is a chronic inflammatory state associated with old age in humans. Periodontitis contributes to inflammaging since periodontitis in the elderly is associated with increased markers of systemic inflammation. Age-related changes also affect neutrophil function, especially antimicrobial activity, so neutrophils may become more pathological. After infiltration into the tissue, neutrophils are equipped with several antimicrobial strategies to reduce the number of antigens. Phagocytosis is the ability of neutrophils to engulf and kill microbes, but neutrophil phagocytosis is weakened in the elderly. Age-related changes affecting neutrophils, macrophages, and T cells appear to promote pathogenic immune responses and contribute to the increased prevalence of periodontal disease in aging individuals. Proper regulation of the host immune response is critical in maintaining periodontal health. This paper aims to describe the aging process and its relation to periodontal conditions.
Periodontal tissue destruction can cause complaints for sufferers. Inflammatory conditions of the gingiva, bleeding gums, and even tooth loss are clinical features of the destruction of the periodontal tissues. Periodontitis is an inflammatory disease involving the periodontal tissues. The prevalence of periodontium destruction increases with aging. Changes in innate and adaptive immunity that occur in the elderly also play a role in the severity of periodontitis. “Inflammaging” is a chronic inflammatory state associated with old age in humans. Periodontitis contributes to inflammaging since periodontitis in the elderly is associated with increased markers of systemic inflammation. Age-related changes also affect neutrophil function, especially antimicrobial activity, so neutrophils may become more pathological. After infiltration into the tissue, neutrophils are equipped with several antimicrobial strategies to reduce the number of antigens. Phagocytosis is the ability of neutrophils to engulf and kill microbes, but neutrophil phagocytosis is weakened in the elderly. Age-related changes affecting neutrophils, macrophages, and T cells appear to promote pathogenic immune responses and contribute to the increased prevalence of periodontal disease in aging individuals. Proper regulation of the host immune response is critical in maintaining periodontal health. This paper aims to describe the aging process and its relation to periodontal conditions.
DOI: https://doi.org/10.37349/ei.2023.00098
This article belongs to the special issue Immunosenescence: Mechanisms and Its Impact

The Src homology 2 (SH2) and SH3 domain-containing chicken tumor virus number 10 (CT10) regulator of kinase (Crk) adaptor proteins include three cellular members that serve as integral constituents of multiple receptor-linked signal transduction pathways. CrkI and CrkII are products of alternative RNA-splicing which is transcribed from a single gene, while Crk-like (CrkL), which is highly homologous to CrkII, is encoded by a different gene. Thanks to their modular structure, the Crk adaptor proteins can simultaneously interact with activated receptors and a wide range of effector molecules, and orchestrate the assembly of complexes containing enzymes and substrates at the receptor site. They are involved in the regulation of a large number of cellular processes which control cell growth, differentiation, transformation, and apoptosis. Cell activation-dependent tyrosine phosphorylation of CrkII and CrkL serves as a major posttranslational modification mechanism that introduces conformational changes in the proteins by promoting an intramolecular interaction between the phosphotyrosine and the self SH2 domain. The resulting conformational change induces downregulation of CrkII- and CrkL-dependent biological processes. A second type of posttranslational modification mechanism regulates the structure and function of the CrkII adaptor protein by immunophilin-mediated protein isomerization. Two of the most abundant immunophilins in T lymphocytes which function as peptidyl-prolyl cis-trans isomerases (PPIases), namely cyclophilin A (CypA) and FK506-binding proteins (FKBPs), can associate with CrkII and catalyze its reciprocal cis-trans isomerization. This mechanism is of special importance for the regulation of T lymphocyte functions and for T cell-mediated immune responses, since immunophilin inhibitors, such as cyclosporin A (CsA) and FK506, function as immunosuppressive drugs that can prevent allotransplanted graft rejection. The present manuscript focuses on selected functions of Crk adaptor proteins, predominantly in T lymphocytes, and reviews in more detail the current knowledge on the immunophilin-dependent regulation of the structure and function of the CrkII adaptor protein.
The Src homology 2 (SH2) and SH3 domain-containing chicken tumor virus number 10 (CT10) regulator of kinase (Crk) adaptor proteins include three cellular members that serve as integral constituents of multiple receptor-linked signal transduction pathways. CrkI and CrkII are products of alternative RNA-splicing which is transcribed from a single gene, while Crk-like (CrkL), which is highly homologous to CrkII, is encoded by a different gene. Thanks to their modular structure, the Crk adaptor proteins can simultaneously interact with activated receptors and a wide range of effector molecules, and orchestrate the assembly of complexes containing enzymes and substrates at the receptor site. They are involved in the regulation of a large number of cellular processes which control cell growth, differentiation, transformation, and apoptosis. Cell activation-dependent tyrosine phosphorylation of CrkII and CrkL serves as a major posttranslational modification mechanism that introduces conformational changes in the proteins by promoting an intramolecular interaction between the phosphotyrosine and the self SH2 domain. The resulting conformational change induces downregulation of CrkII- and CrkL-dependent biological processes. A second type of posttranslational modification mechanism regulates the structure and function of the CrkII adaptor protein by immunophilin-mediated protein isomerization. Two of the most abundant immunophilins in T lymphocytes which function as peptidyl-prolyl cis-trans isomerases (PPIases), namely cyclophilin A (CypA) and FK506-binding proteins (FKBPs), can associate with CrkII and catalyze its reciprocal cis-trans isomerization. This mechanism is of special importance for the regulation of T lymphocyte functions and for T cell-mediated immune responses, since immunophilin inhibitors, such as cyclosporin A (CsA) and FK506, function as immunosuppressive drugs that can prevent allotransplanted graft rejection. The present manuscript focuses on selected functions of Crk adaptor proteins, predominantly in T lymphocytes, and reviews in more detail the current knowledge on the immunophilin-dependent regulation of the structure and function of the CrkII adaptor protein.
DOI: https://doi.org/10.37349/ei.2023.00099
This article belongs to the special issue The Role of Adaptor Proteins in Lymphoid Cell Signaling

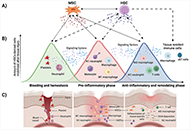
Besides trauma, several pathological conditions which directly affect the normal functioning of organs, require new therapeutic strategies to repair damaged or diseased tissues. Tissue regeneration is a complex and spatiotemporal process involving a plethora of cell types, including various immune cells and stem cells in a synchronized relationship. However, individual parameters, namely ageing, obesity, diabetes, and chronic conditions, have been intrinsically correlated with poor regenerative properties of adult tissues. While vast progress has been made regarding stem cell-based therapy to direct self-healing, the immune response is still the Achilles’ heel of such strategies. Whereas the role of effector immune cells has been well defined along the regenerative process, an understanding of the behavior of the main adult stem cells, namely mesenchymal stem cells (MSCs) and hematopoietic stem and progenitor cells (HSPCs), along the different phases of the regenerative process could clarify how these stem cells can be used to positively influence the immune response. In this scope, this review highlights the main interactions between these stem cells and immune cells during tissue repair, exploring the most important regenerative properties of stem cells and correlating them with the modulation of the immune response during tissue regeneration. Furthermore, the utmost strategies used to explore how the behavior and stem cell fate are affected by specific microenvironments and/or stimuli usually found during a regenerative process, are emphasized. This clarification may provide critical insight into the molecular mechanisms by which stem cells modulate the immune response in a positive feedback loop toward tissue repair.
Besides trauma, several pathological conditions which directly affect the normal functioning of organs, require new therapeutic strategies to repair damaged or diseased tissues. Tissue regeneration is a complex and spatiotemporal process involving a plethora of cell types, including various immune cells and stem cells in a synchronized relationship. However, individual parameters, namely ageing, obesity, diabetes, and chronic conditions, have been intrinsically correlated with poor regenerative properties of adult tissues. While vast progress has been made regarding stem cell-based therapy to direct self-healing, the immune response is still the Achilles’ heel of such strategies. Whereas the role of effector immune cells has been well defined along the regenerative process, an understanding of the behavior of the main adult stem cells, namely mesenchymal stem cells (MSCs) and hematopoietic stem and progenitor cells (HSPCs), along the different phases of the regenerative process could clarify how these stem cells can be used to positively influence the immune response. In this scope, this review highlights the main interactions between these stem cells and immune cells during tissue repair, exploring the most important regenerative properties of stem cells and correlating them with the modulation of the immune response during tissue regeneration. Furthermore, the utmost strategies used to explore how the behavior and stem cell fate are affected by specific microenvironments and/or stimuli usually found during a regenerative process, are emphasized. This clarification may provide critical insight into the molecular mechanisms by which stem cells modulate the immune response in a positive feedback loop toward tissue repair.
DOI: https://doi.org/10.37349/ei.2023.00100

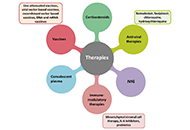
The coronavirus disease-2019 (COVID-19) pandemic is a significant threat in the modern era. Clinical studies show that the most common symptom of severe COVID-19 is viral pneumonia-induced acute respiratory distress syndrome (ARDS). The underlying mechanisms by which severe respiratory disease syndrome-coronavirus-2 (SARS-CoV-2) results in ARDS and how certain host factors confer an increased risk of developing severe disease remain unknown. Therefore, identifying the distinctive features of this severe and fatal disease and the therapeutic approaches to COVID-19-induced ARDS remains an immediate need to serve as a basis for best practice models of standardized ARDS treatment. This review article aims to comprehensively discuss the immunopathology of ARDS and provides an overview of the precise role of both the innate and adaptive immune system, with emphasis on the current treatment strategies being tested in the COVID-19-induced ARDS patients. This knowledge will supposedly help in revealing further mechanistic insights into understanding COVID-19-induced ARDS.
The coronavirus disease-2019 (COVID-19) pandemic is a significant threat in the modern era. Clinical studies show that the most common symptom of severe COVID-19 is viral pneumonia-induced acute respiratory distress syndrome (ARDS). The underlying mechanisms by which severe respiratory disease syndrome-coronavirus-2 (SARS-CoV-2) results in ARDS and how certain host factors confer an increased risk of developing severe disease remain unknown. Therefore, identifying the distinctive features of this severe and fatal disease and the therapeutic approaches to COVID-19-induced ARDS remains an immediate need to serve as a basis for best practice models of standardized ARDS treatment. This review article aims to comprehensively discuss the immunopathology of ARDS and provides an overview of the precise role of both the innate and adaptive immune system, with emphasis on the current treatment strategies being tested in the COVID-19-induced ARDS patients. This knowledge will supposedly help in revealing further mechanistic insights into understanding COVID-19-induced ARDS.
DOI: https://doi.org/10.37349/ei.2023.00101
This article belongs to the special issue Immunology, Immunopathology and Genomics of SARS-COV-2

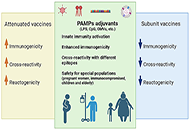
The emergence and re-emergence of pathogens is a public-health concern, which has become more evident after the coronavirus disease 2019 (COVID-19) pandemic and the monkeypox outbreaks in early 2022. Given that vaccines are the more effective and affordable tools to control infectious diseases, the authors reviewed two heterologous effects of vaccines: the trained immunity and the cross-reactivity. Trained immunity, provided by attenuated vaccines, was exemplified in this article by the decreased the burden of COVID-19 in populations with high Bacille Calmette-Guerin (BCG) coverage. Cross-reactive responses were exemplified here by the studies which suggested that vaccinia could help controlling the monkeypox outbreak, because of common epitopes shared by orthopoxviruses. Although modern vaccination is likely to use subunit vaccines, the authors discussed how adjuvants might be the key to induce trained immunity and improve cross-reactive responses, ensuring that heterologous effects would improve the vaccine’s response.
The emergence and re-emergence of pathogens is a public-health concern, which has become more evident after the coronavirus disease 2019 (COVID-19) pandemic and the monkeypox outbreaks in early 2022. Given that vaccines are the more effective and affordable tools to control infectious diseases, the authors reviewed two heterologous effects of vaccines: the trained immunity and the cross-reactivity. Trained immunity, provided by attenuated vaccines, was exemplified in this article by the decreased the burden of COVID-19 in populations with high Bacille Calmette-Guerin (BCG) coverage. Cross-reactive responses were exemplified here by the studies which suggested that vaccinia could help controlling the monkeypox outbreak, because of common epitopes shared by orthopoxviruses. Although modern vaccination is likely to use subunit vaccines, the authors discussed how adjuvants might be the key to induce trained immunity and improve cross-reactive responses, ensuring that heterologous effects would improve the vaccine’s response.
DOI: https://doi.org/10.37349/ei.2023.00102

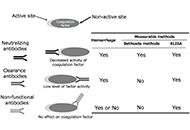
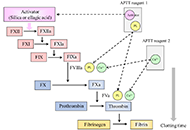
In patients with autoimmune coagulation factor deficiency (AiCFD), the production of autoantibodies that inhibit coagulation factors in the blood reduces the activity of those relevant coagulation factors, resulting in severe bleeding symptoms. Recently, reports of patients with AiCFD have noted the concomitant detection of lupus anticoagulant (LA), a risk factor for thrombosis. LA-positive patients may show bleeding symptoms due to decreased activity of coagulation factor II (FII) caused by autoantibodies against FII, in addition to thrombotic symptoms, a condition termed LA-hypoprothrombinemia syndrome (LAHPS). Anti-FII antibodies in LAHPS cases are frequently cleared antibodies that can be detected using immunological techniques, such as enzyme-linked immunosorbent assay (ELISA). Recently, several cases of coagulation FV inhibitors, known as autoimmune FV deficiency, have been reported. Some of these cases may be complicated by LA, which can cause thrombosis. False-positive results for anticoagulant inhibitors are known to occur in LA cases; therefore, immunological confirmation of antibodies against coagulation factors is recommended. Additionally, acquired hemophilia A (AHA), caused by autoantibodies against FVIII, is a typical acquired hemorrhagic diathesis, although affected patients may present with thrombosis associated with LA. Thus, it is important to remember that hemorrhagic diathesis due to autoantibodies against clotting factors can also result in thrombosis, as demonstrated by the co-detection of LA. When clotting factor inhibitors are detected in LA-positive individuals, it is important to confirm the presence of autoantibodies against coagulation factors using immunological methods, such as ELISA, to avoid false-positive results.
In patients with autoimmune coagulation factor deficiency (AiCFD), the production of autoantibodies that inhibit coagulation factors in the blood reduces the activity of those relevant coagulation factors, resulting in severe bleeding symptoms. Recently, reports of patients with AiCFD have noted the concomitant detection of lupus anticoagulant (LA), a risk factor for thrombosis. LA-positive patients may show bleeding symptoms due to decreased activity of coagulation factor II (FII) caused by autoantibodies against FII, in addition to thrombotic symptoms, a condition termed LA-hypoprothrombinemia syndrome (LAHPS). Anti-FII antibodies in LAHPS cases are frequently cleared antibodies that can be detected using immunological techniques, such as enzyme-linked immunosorbent assay (ELISA). Recently, several cases of coagulation FV inhibitors, known as autoimmune FV deficiency, have been reported. Some of these cases may be complicated by LA, which can cause thrombosis. False-positive results for anticoagulant inhibitors are known to occur in LA cases; therefore, immunological confirmation of antibodies against coagulation factors is recommended. Additionally, acquired hemophilia A (AHA), caused by autoantibodies against FVIII, is a typical acquired hemorrhagic diathesis, although affected patients may present with thrombosis associated with LA. Thus, it is important to remember that hemorrhagic diathesis due to autoantibodies against clotting factors can also result in thrombosis, as demonstrated by the co-detection of LA. When clotting factor inhibitors are detected in LA-positive individuals, it is important to confirm the presence of autoantibodies against coagulation factors using immunological methods, such as ELISA, to avoid false-positive results.
DOI: https://doi.org/10.37349/ei.2023.00103
This article belongs to the special issue Autoantibodies Associated to Thrombosis and Hemostasis

Changes occurring in the immune system along the ageing process increase the risk of infection, susceptibility to tumor development, and autoimmunity. Interventions such as physical exercise, supplements, and probiotics have been proposed in order to circumvent these conditions. Vitamin D supplementation could contribute to the immune system homeostasis in older adults since a large proportion of this population has low levels of circulating vitamin D. Additionally, observational studies have shown the association between vitamin D status and infections, chronic diseases such as cancer, diabetes, and cardiovascular disease. Recently it was observed that old patients with COVID-19 and vitamin D deficiency had enhanced severity of lung damage, longer stay at the hospital, and increased risk of death, suggesting that vitamin D plays an important role in the patient outcome from COVID-19. A high dose of vitamin D supplementation improved clinical recovery in a case-series report but in another study, no evident link between levels of vitamin D and risk of COVID-19 infection was found. Results also remain debatable for vitamin D supplements and improvement of immune response after vaccination, tuberculosis, pneumonia, and sepsis. It has been hypothesized that vitamin D could modulate the immune system and thus provide both efficacies in the immune response to pathogens/vaccinations and reduction of the inflammatory phenotype. This review will discuss vitamin D and homeostasis of the immune system; the literature-based clinical data on vitamin D and infections; and the possible link between vitamin D and immune response after vaccination.
Changes occurring in the immune system along the ageing process increase the risk of infection, susceptibility to tumor development, and autoimmunity. Interventions such as physical exercise, supplements, and probiotics have been proposed in order to circumvent these conditions. Vitamin D supplementation could contribute to the immune system homeostasis in older adults since a large proportion of this population has low levels of circulating vitamin D. Additionally, observational studies have shown the association between vitamin D status and infections, chronic diseases such as cancer, diabetes, and cardiovascular disease. Recently it was observed that old patients with COVID-19 and vitamin D deficiency had enhanced severity of lung damage, longer stay at the hospital, and increased risk of death, suggesting that vitamin D plays an important role in the patient outcome from COVID-19. A high dose of vitamin D supplementation improved clinical recovery in a case-series report but in another study, no evident link between levels of vitamin D and risk of COVID-19 infection was found. Results also remain debatable for vitamin D supplements and improvement of immune response after vaccination, tuberculosis, pneumonia, and sepsis. It has been hypothesized that vitamin D could modulate the immune system and thus provide both efficacies in the immune response to pathogens/vaccinations and reduction of the inflammatory phenotype. This review will discuss vitamin D and homeostasis of the immune system; the literature-based clinical data on vitamin D and infections; and the possible link between vitamin D and immune response after vaccination.
DOI: https://doi.org/10.37349/ei.2023.00106
This article belongs to the special issue Immunosenescence: Mechanisms and Its Impact

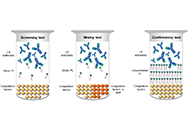
Accurate lupus anticoagulant (LA) detection is crucial to antiphospholipid syndrome (APS) diagnosis. Detection is based on LA functional behavior in coagulation assays irrespective of epitope specificity. LA screening tests employ dilute phospholipids to accentuate in vitro inhibition by LAs, although they are not LA-specific and can be elevated by other coagulation abnormalities. Elevated screening tests are reflexed to mixing tests to distinguish between factor deficiency and inhibition. Confirmatory tests with high phospholipid concentration swamp LA to generate shorter clotting times than screening tests, whilst prolongation persists with non-phospholipid-dependent inhibitors. LA heterogeneity means that no single screening test detects every LA and the screen/mix/confirm medley must be applied to at least two assay types, usually dilute Russell’s viper venom time (dRVVT) and an LA-sensitive activated partial thromboplastin time (aPTT). Most laboratories restrict LA testing to these two assays, yet others, such as dilute prothrombin time (dPT), can perform with equal diagnostic efficacy, and additionally detect LA unreactive with dRVVT and aPTT. Converting clotting times to normalized ratios improves assay performance, and practitioners must choose between normal pooled plasma (NPP) clotting time denominators to reflect on-the-day assay performance, or reference interval (RI) mean clotting times to negate the effects of NPP variation. Cut-offs can be generated parametrically from normally distributed data, or different percentiles applied depending on the preferred balance between sensitivity and specificity. Sourcing sufficient donors for accurate cut-off estimations is problematic and transference exercises can be undertaken on low donor numbers. Analytical limitations of mixing tests have led to the adoption of alternative algorithms to the screen/mix/confirm test order, whilst some continue to rigidly apply the latter despite those limitations. Strategies to reduce or eliminate the effects of therapeutic anticoagulation have limitations, whilst the Taipan snake venom time (TSVT) screening test with an ecarin time (ET) confirmatory test is insensitive to vitamin K antagonist (VKA) and direct activated factor X anticoagulation.
Accurate lupus anticoagulant (LA) detection is crucial to antiphospholipid syndrome (APS) diagnosis. Detection is based on LA functional behavior in coagulation assays irrespective of epitope specificity. LA screening tests employ dilute phospholipids to accentuate in vitro inhibition by LAs, although they are not LA-specific and can be elevated by other coagulation abnormalities. Elevated screening tests are reflexed to mixing tests to distinguish between factor deficiency and inhibition. Confirmatory tests with high phospholipid concentration swamp LA to generate shorter clotting times than screening tests, whilst prolongation persists with non-phospholipid-dependent inhibitors. LA heterogeneity means that no single screening test detects every LA and the screen/mix/confirm medley must be applied to at least two assay types, usually dilute Russell’s viper venom time (dRVVT) and an LA-sensitive activated partial thromboplastin time (aPTT). Most laboratories restrict LA testing to these two assays, yet others, such as dilute prothrombin time (dPT), can perform with equal diagnostic efficacy, and additionally detect LA unreactive with dRVVT and aPTT. Converting clotting times to normalized ratios improves assay performance, and practitioners must choose between normal pooled plasma (NPP) clotting time denominators to reflect on-the-day assay performance, or reference interval (RI) mean clotting times to negate the effects of NPP variation. Cut-offs can be generated parametrically from normally distributed data, or different percentiles applied depending on the preferred balance between sensitivity and specificity. Sourcing sufficient donors for accurate cut-off estimations is problematic and transference exercises can be undertaken on low donor numbers. Analytical limitations of mixing tests have led to the adoption of alternative algorithms to the screen/mix/confirm test order, whilst some continue to rigidly apply the latter despite those limitations. Strategies to reduce or eliminate the effects of therapeutic anticoagulation have limitations, whilst the Taipan snake venom time (TSVT) screening test with an ecarin time (ET) confirmatory test is insensitive to vitamin K antagonist (VKA) and direct activated factor X anticoagulation.
DOI: https://doi.org/10.37349/ei.2023.00104
This article belongs to the special issue Autoantibodies Associated to Thrombosis and Hemostasis

Aim:
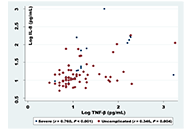
A number of questions remain unanswered concerning how infected individuals regulate their immune response to Plasmodium falciparum (P. falciparum) parasites at varying levels of exposure. Due to the interactions of inflammatory mediators and cytokines with the P. falciparum parasite complex density, several mediators influence parasitaemia and may give some indications of disease severity and represent effective signs in clinical manifestations of malaria disease.
Methods:
In this study, various levels of immune response mediators of interleukin 8 (IL-8), tumor necrosis factor-beta (TNF-β, also known as lymphotoxin-α), interferon-gamma (IFN-γ), IL-6, and IL-10 were investigated to the different phases of infection with P. falciparum in hyperendemic states in Sudan (White Nile, Blue Nile). This study vetted the association between certain inflammatory mediators during malaria infection and parasite density. This study was based on a total of 108 cases, in which 86 patients (62.0%) were uncomplicated and (17.6%) were severe, all met the diagnostic criteria and were clinically admitted for malaria infections. Commercial enzyme-linked immunosorbent assay (ELISA) kits were employed to determine the inflammatory mediator’s serum concentration.
Results:
The analysis of data indicated that older infected children had substantially raised levels of IFN-γ (P < 0.05), among study groups, levels of IFN-γ, TNF-β, and IL-8 were strongly linked with the severity of malaria, in severe and uncomplicated cases (P < 0.001), IL-6 and IL-10 were significantly associated with severe malaria cases uniquely (P < 0.001). Furthermore, we reported a positive correlation between IL-8 and TNF-β during all infection cases (r = 0.760, P < 0.001). Additionally, in severe malaria cases IL-6 was positively correlated with IL-10 (r = 0.575, P = 0.010).
Conclusions:
Eliminating P. falciparum blood-stage infection needs effective, specific, and tuned immune response strategies, which may present in the mediator’s correlations and depend on the density of the infection. Besides the effective levels contribution of certain cytokines that play protective roles during different stages of an infection.
Aim:
A number of questions remain unanswered concerning how infected individuals regulate their immune response to Plasmodium falciparum (P. falciparum) parasites at varying levels of exposure. Due to the interactions of inflammatory mediators and cytokines with the P. falciparum parasite complex density, several mediators influence parasitaemia and may give some indications of disease severity and represent effective signs in clinical manifestations of malaria disease.
Methods:
In this study, various levels of immune response mediators of interleukin 8 (IL-8), tumor necrosis factor-beta (TNF-β, also known as lymphotoxin-α), interferon-gamma (IFN-γ), IL-6, and IL-10 were investigated to the different phases of infection with P. falciparum in hyperendemic states in Sudan (White Nile, Blue Nile). This study vetted the association between certain inflammatory mediators during malaria infection and parasite density. This study was based on a total of 108 cases, in which 86 patients (62.0%) were uncomplicated and (17.6%) were severe, all met the diagnostic criteria and were clinically admitted for malaria infections. Commercial enzyme-linked immunosorbent assay (ELISA) kits were employed to determine the inflammatory mediator’s serum concentration.
Results:
The analysis of data indicated that older infected children had substantially raised levels of IFN-γ (P < 0.05), among study groups, levels of IFN-γ, TNF-β, and IL-8 were strongly linked with the severity of malaria, in severe and uncomplicated cases (P < 0.001), IL-6 and IL-10 were significantly associated with severe malaria cases uniquely (P < 0.001). Furthermore, we reported a positive correlation between IL-8 and TNF-β during all infection cases (r = 0.760, P < 0.001). Additionally, in severe malaria cases IL-6 was positively correlated with IL-10 (r = 0.575, P = 0.010).
Conclusions:
Eliminating P. falciparum blood-stage infection needs effective, specific, and tuned immune response strategies, which may present in the mediator’s correlations and depend on the density of the infection. Besides the effective levels contribution of certain cytokines that play protective roles during different stages of an infection.
DOI: https://doi.org/10.37349/ei.2023.00109

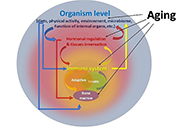
Interest in the mechanisms of aging of the immune system has not faded over the past 100 years, and it is caused by the immune-mediated development of age-related pathology, including autoimmune organ damage, reduced vaccination efficiency, atherosclerosis, the development of cardiovascular pathology, etc. In contrast to many other organs and systems, the immune system aging begins at an early age and has more pronounced changes that lead to the development of secondary pathology, which significantly affects life expectancy. But an effective strategy to restore immune function has not been developed yet. During this time, the mechanisms of age-related dysfunction of organs and cells of both the adaptive and innate immune systems were studied in detail—thymus involution, a decrease in the potential of hematopoietic stem cells, impaired differentiation and functions of immunocompetent cells, as well as the ways of their interaction. Numerous potential therapeutic targets have been identified and various approaches have been used to implement such therapeutic interventions. The review is devoted to replacement therapy using transplantation of hematopoietic stem cells (HSCs) and young lymphoid cells and tissues, cellular and systemic factor exchange in heterochronic parabiosis, and some other widely used life extension approaches. It has been proven that cell therapy using young cells to rejuvenate the old immune system, unfortunately, often turns out to be ineffective because it does not eliminate the root cause of age-related changes. The phenomenon of inflamm-aging that develops with age can significantly affect both the aging of the organism in general and the functioning of immunocompetent cells in particular. Therefore, the most promising direction in the restoration of immune functions during aging is systemic approaches that have a complex effect on the organism as a whole and can slow down the aging process.
Interest in the mechanisms of aging of the immune system has not faded over the past 100 years, and it is caused by the immune-mediated development of age-related pathology, including autoimmune organ damage, reduced vaccination efficiency, atherosclerosis, the development of cardiovascular pathology, etc. In contrast to many other organs and systems, the immune system aging begins at an early age and has more pronounced changes that lead to the development of secondary pathology, which significantly affects life expectancy. But an effective strategy to restore immune function has not been developed yet. During this time, the mechanisms of age-related dysfunction of organs and cells of both the adaptive and innate immune systems were studied in detail—thymus involution, a decrease in the potential of hematopoietic stem cells, impaired differentiation and functions of immunocompetent cells, as well as the ways of their interaction. Numerous potential therapeutic targets have been identified and various approaches have been used to implement such therapeutic interventions. The review is devoted to replacement therapy using transplantation of hematopoietic stem cells (HSCs) and young lymphoid cells and tissues, cellular and systemic factor exchange in heterochronic parabiosis, and some other widely used life extension approaches. It has been proven that cell therapy using young cells to rejuvenate the old immune system, unfortunately, often turns out to be ineffective because it does not eliminate the root cause of age-related changes. The phenomenon of inflamm-aging that develops with age can significantly affect both the aging of the organism in general and the functioning of immunocompetent cells in particular. Therefore, the most promising direction in the restoration of immune functions during aging is systemic approaches that have a complex effect on the organism as a whole and can slow down the aging process.
DOI: https://doi.org/10.37349/ei.2023.00105
This article belongs to the special issue Immunosenescence: Mechanisms and Its Impact

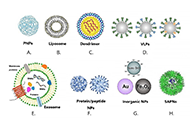
Vaccination has made an enormous contribution to global health. Treatment resistance for infectious diseases is growing quickly, and chemotherapeutic toxicity in cancer means that vaccines must be made right away to save humanity. But subunit vaccinations alone don’t give enough strong and long-lasting protection against infections that can kill. Nanoparticle (NP)-based delivery vehicles, such as dendrimers, liposomes, micelles, virosomes, nanogels, and microemulsions, offer interesting ways to get around the problems with traditional vaccine adjuvants. The nanovaccines (50–250 nm in size) are most efficient in terms of tissue targeting, staying in the bloodstream for a long time. Nanovaccines can improve antigen presentation, targeted delivery, stimulation of the body’s innate immune system, and a strong T-cell response without putting people at risk. This can help fight infectious diseases and cancers. Also, nanovaccines can be very helpful for making cancer treatments that use immunotherapy. So, this review highlights the various types of NPs used in the techniques that have worked in the new paradigm in viral vaccinology for infectious diseases. It gives a full rundown of the current NP-based vaccines, their potential as adjuvants, and the ways they can be delivered to cells. In the future, the best nanovaccines will try to be more logically designed, have more antigens in them, be fully functionalized, and be given to the right people.
Vaccination has made an enormous contribution to global health. Treatment resistance for infectious diseases is growing quickly, and chemotherapeutic toxicity in cancer means that vaccines must be made right away to save humanity. But subunit vaccinations alone don’t give enough strong and long-lasting protection against infections that can kill. Nanoparticle (NP)-based delivery vehicles, such as dendrimers, liposomes, micelles, virosomes, nanogels, and microemulsions, offer interesting ways to get around the problems with traditional vaccine adjuvants. The nanovaccines (50–250 nm in size) are most efficient in terms of tissue targeting, staying in the bloodstream for a long time. Nanovaccines can improve antigen presentation, targeted delivery, stimulation of the body’s innate immune system, and a strong T-cell response without putting people at risk. This can help fight infectious diseases and cancers. Also, nanovaccines can be very helpful for making cancer treatments that use immunotherapy. So, this review highlights the various types of NPs used in the techniques that have worked in the new paradigm in viral vaccinology for infectious diseases. It gives a full rundown of the current NP-based vaccines, their potential as adjuvants, and the ways they can be delivered to cells. In the future, the best nanovaccines will try to be more logically designed, have more antigens in them, be fully functionalized, and be given to the right people.
DOI: https://doi.org/10.37349/ei.2023.00107
This article belongs to the special issue Old and New Paradigms in Viral Vaccinology

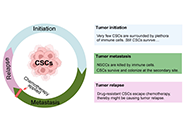
Cancer stem cells (CSCs) are a small subpopulation of cells that drive the formation and progression of tumors. However, during tumor initiation, how CSCs communicate with neighbouring immune cells to overcome the powerful immune surveillance barrier in order to form, spread, and maintain the tumor, remains poorly understood. It is, therefore, absolutely necessary to understand how a small number of tumor-initiating cells (TICs) survive immune attack during (a) the “elimination phase” of “tumor immune-editing”, (b) the establishment of regional or distant tumor after metastasis, and (c) recurrence after therapy. Mounting evidence suggests that CSCs suppress the immune system through a variety of distinct mechanisms that ensure the survival of not only CSCs but also non-stem cancer cells (NSCCs), which eventually form the tumor mass. In this review article, the mechanisms via which CSCs change the immune landscape of the tissue of origin, which contains macrophages, dendritic cells (DCs), myeloid-derived suppressor cells (MDSCs), natural killer (NK) cells, and tumor-infiltrating lymphocytes, in favour of tumorigenesis were discussed. The failure of cancer immunotherapy might also be explained by such interaction between CSCs and immune cells. This review will shed light on the critical role of CSCs in tumor immune evasion and emphasize the importance of CSC-targeted immunotherapy as a cutting-edge technique for battling cancer by restricting communication between immune cells and CSCs.
Cancer stem cells (CSCs) are a small subpopulation of cells that drive the formation and progression of tumors. However, during tumor initiation, how CSCs communicate with neighbouring immune cells to overcome the powerful immune surveillance barrier in order to form, spread, and maintain the tumor, remains poorly understood. It is, therefore, absolutely necessary to understand how a small number of tumor-initiating cells (TICs) survive immune attack during (a) the “elimination phase” of “tumor immune-editing”, (b) the establishment of regional or distant tumor after metastasis, and (c) recurrence after therapy. Mounting evidence suggests that CSCs suppress the immune system through a variety of distinct mechanisms that ensure the survival of not only CSCs but also non-stem cancer cells (NSCCs), which eventually form the tumor mass. In this review article, the mechanisms via which CSCs change the immune landscape of the tissue of origin, which contains macrophages, dendritic cells (DCs), myeloid-derived suppressor cells (MDSCs), natural killer (NK) cells, and tumor-infiltrating lymphocytes, in favour of tumorigenesis were discussed. The failure of cancer immunotherapy might also be explained by such interaction between CSCs and immune cells. This review will shed light on the critical role of CSCs in tumor immune evasion and emphasize the importance of CSC-targeted immunotherapy as a cutting-edge technique for battling cancer by restricting communication between immune cells and CSCs.
DOI: https://doi.org/10.37349/ei.2023.00108

Antiphospholipid syndrome (APS) is defined as an autoimmune and prothrombotic disorder in patients with the persistent presence of antiphospholipid antibodies (aPLs). In the classification criteria, aPL expresses lupus anticoagulant (LA) activity, which is detected by prolongation of coagulation assays. The LA detection algorithm is a sequential flow including screening tests, mixing tests, and confirmatory tests to differentiate between LA-positive and other anticoagulant abnormalities. Two types of assays are used, like dilute Russell’s viper venom time (dRVVT) and activated partial thromboplastin time (APTT) because no single test is sensitive to all LAs. The anticoagulant drugs prescribed for the prevention and treatment of thrombosis disorders can interfere with the assays, and it is important to know the effects of these drugs in the assays. Especially, new generation anticoagulant drugs, called direct oral anticoagulants (DOACs), affect the results. In this review, the following points are discussed: i) LA detection flow and data interpretation, ii) the principles of coagulation assays proposed and their characteristics, and iii) the effects of anticoagulant drugs in LA detection.
Antiphospholipid syndrome (APS) is defined as an autoimmune and prothrombotic disorder in patients with the persistent presence of antiphospholipid antibodies (aPLs). In the classification criteria, aPL expresses lupus anticoagulant (LA) activity, which is detected by prolongation of coagulation assays. The LA detection algorithm is a sequential flow including screening tests, mixing tests, and confirmatory tests to differentiate between LA-positive and other anticoagulant abnormalities. Two types of assays are used, like dilute Russell’s viper venom time (dRVVT) and activated partial thromboplastin time (APTT) because no single test is sensitive to all LAs. The anticoagulant drugs prescribed for the prevention and treatment of thrombosis disorders can interfere with the assays, and it is important to know the effects of these drugs in the assays. Especially, new generation anticoagulant drugs, called direct oral anticoagulants (DOACs), affect the results. In this review, the following points are discussed: i) LA detection flow and data interpretation, ii) the principles of coagulation assays proposed and their characteristics, and iii) the effects of anticoagulant drugs in LA detection.
DOI: https://doi.org/10.37349/ei.2023.00110
This article belongs to the special issue Autoantibodies Associated to Thrombosis and Hemostasis

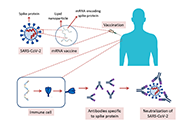
Vaccines are prophylactic medical products effectively used against infectious diseases. Although a high amount of vaccine studies are conducted at the preclinical stage, the number of approved vaccines is less than 10%. Development of vaccines from the research stage to the approval of administrative institutions takes about 5 years to 10 years conventionally. However, this period of time for vaccine development is not convenient during public health emergencies because an effective vaccine is required in a short time to restrict the speed of high mortality and morbidity. The pandemic of coronavirus disease 2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), had its catastrophic effects worldwide quickly. Therefore, an atypical process was followed for the development of COVID-19 vaccines. Great effort was spent in terms of cooperation among the governmental institutions, academia, and medical companies as well as a high amount of budget was allocated to develop effective vaccines against COVID-19. As of March 2023, the numbers of COVID-19 vaccines in clinical and preclinical development were 183 and 199, respectively. An emergency use authorization (EUA) process was applied to accelerate the approval of the vaccines. Consequently, vaccinations could be started in less than a year, which decelerated the speed of the pandemic. Although EUA caused hesitancy among some people questioning the safety and efficacy of the vaccines, the vast majority of the population was vaccinated. Currently, more than 5.5 billion people (about 70% of the world population) have received 13.38 billion doses of 11 different COVID-19 vaccines, and 73% of the doses were Comirnaty manufactured by Pfizer/BioNTech.
Vaccines are prophylactic medical products effectively used against infectious diseases. Although a high amount of vaccine studies are conducted at the preclinical stage, the number of approved vaccines is less than 10%. Development of vaccines from the research stage to the approval of administrative institutions takes about 5 years to 10 years conventionally. However, this period of time for vaccine development is not convenient during public health emergencies because an effective vaccine is required in a short time to restrict the speed of high mortality and morbidity. The pandemic of coronavirus disease 2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), had its catastrophic effects worldwide quickly. Therefore, an atypical process was followed for the development of COVID-19 vaccines. Great effort was spent in terms of cooperation among the governmental institutions, academia, and medical companies as well as a high amount of budget was allocated to develop effective vaccines against COVID-19. As of March 2023, the numbers of COVID-19 vaccines in clinical and preclinical development were 183 and 199, respectively. An emergency use authorization (EUA) process was applied to accelerate the approval of the vaccines. Consequently, vaccinations could be started in less than a year, which decelerated the speed of the pandemic. Although EUA caused hesitancy among some people questioning the safety and efficacy of the vaccines, the vast majority of the population was vaccinated. Currently, more than 5.5 billion people (about 70% of the world population) have received 13.38 billion doses of 11 different COVID-19 vaccines, and 73% of the doses were Comirnaty manufactured by Pfizer/BioNTech.
DOI: https://doi.org/10.37349/ei.2023.00111
This article belongs to the special issue Old and New Paradigms in Viral Vaccinology

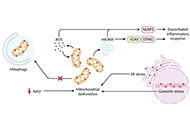
Immunosenescence encompasses multiple age-related adaptations that result in increased susceptibility to infections, chronic inflammatory disorders, and higher mortality risk. Macrophages are key innate cells implicated in inflammatory responses and tissue homeostasis, functions progressively compromised by aging. This process coincides with declining mitochondrial physiology, whose integrity is required to sustain and orchestrate immune responses. Indeed, multiple insults observed in aged macrophages have been implied as drivers of mitochondrial dysfunction, but how this translates into impaired immune function remains sparsely explored. This review provides a perspective on recent studies elucidating the underlying mechanisms linking dysregulated mitochondria homeostasis to immune function in aged macrophages. Genomic stress alongside defective mitochondrial turnover accounted for the progressive accumulation of damaged mitochondria in aged macrophages, thus resulting in a higher susceptibility to excessive mitochondrial DNA (mtDNA) leakage and reactive oxygen species (ROS) production. Increased levels of these mitochondrial products following infection were demonstrated to contribute to exacerbated inflammatory responses mediated by overstimulation of NOD-like receptor family pyrin domain containing 3 (NLRP3) inflammasome and cyclic GMP-ATP synthase (cGAS)-stimulator of interferon genes (STING) pathways. While these mechanisms are not fully elucidated, the present evidence provides a promising area to be explored and a renewed perspective of potential therapeutic targets for immunological dysfunction.
Immunosenescence encompasses multiple age-related adaptations that result in increased susceptibility to infections, chronic inflammatory disorders, and higher mortality risk. Macrophages are key innate cells implicated in inflammatory responses and tissue homeostasis, functions progressively compromised by aging. This process coincides with declining mitochondrial physiology, whose integrity is required to sustain and orchestrate immune responses. Indeed, multiple insults observed in aged macrophages have been implied as drivers of mitochondrial dysfunction, but how this translates into impaired immune function remains sparsely explored. This review provides a perspective on recent studies elucidating the underlying mechanisms linking dysregulated mitochondria homeostasis to immune function in aged macrophages. Genomic stress alongside defective mitochondrial turnover accounted for the progressive accumulation of damaged mitochondria in aged macrophages, thus resulting in a higher susceptibility to excessive mitochondrial DNA (mtDNA) leakage and reactive oxygen species (ROS) production. Increased levels of these mitochondrial products following infection were demonstrated to contribute to exacerbated inflammatory responses mediated by overstimulation of NOD-like receptor family pyrin domain containing 3 (NLRP3) inflammasome and cyclic GMP-ATP synthase (cGAS)-stimulator of interferon genes (STING) pathways. While these mechanisms are not fully elucidated, the present evidence provides a promising area to be explored and a renewed perspective of potential therapeutic targets for immunological dysfunction.
DOI: https://doi.org/10.37349/ei.2023.00112
This article belongs to the special issue Immunosenescence: Mechanisms and Its Impact

