Affiliation:
1Product Development Department, Product Designing 1st section, Fujirebio Inc., Tokyo 107-0052, Japan
2Product Development Department, Fujirebio Europe N.V., 9052 Gent, Belgium
†These authors contributed equally to this work.
Affiliation:
2Product Development Department, Fujirebio Europe N.V., 9052 Gent, Belgium
†These authors contributed equally to this work.
Affiliation:
2Product Development Department, Fujirebio Europe N.V., 9052 Gent, Belgium
ORCID: https://orcid.org/0009-0005-2996-6698
Affiliation:
3Fundamental Research Department, Assay Technology Research Section, Fujirebio Inc., Tokyo 107-0052, Japan
Affiliation:
1Product Development Department, Product Designing 1st section, Fujirebio Inc., Tokyo 107-0052, Japan
Affiliation:
1Product Development Department, Product Designing 1st section, Fujirebio Inc., Tokyo 107-0052, Japan
Affiliation:
4Research and Development Division, Fujirebio Inc., Tokyo 107-0052, Japan
ORCID: https://orcid.org/0000-0003-3818-7985
Affiliation:
1Product Development Department, Product Designing 1st section, Fujirebio Inc., Tokyo 107-0052, Japan
†These authors contributed equally to this work.
Email: hisashi.nojima@hugp.com
ORCID: https://orcid.org/0000-0002-8907-5767
Explor Neurosci. 2023;2:238–244 DOl: https://doi.org/10.37349/en.2023.00024
Received: June 12, 2023 Accepted: August 31, 2023 Published: October 16, 2023
Academic Editor: Dirk M. Hermann, University of Duisburg-Essen, Germany
The article belongs to the special issue Alzheimer’s Disease
Aim: Apolipoprotein E (ApoE) isoforms, especially the ApoE4 isoform, are genetic risk factors for Alzheimer’s disease (AD). Moreover, the APOE ε4 haplotype has a dose-dependent association with an increased risk of amyloid-related imaging abnormalities (ARIA) in individuals receiving disease-modifying therapy for AD. Therefore, the importance of APOE genotyping or proteotyping has been highlighted. Here, the authors developed fully automated chemiluminescence enzyme-immunoassay kit for ApoE4 and Pan-ApoE, and evaluated their diagnostic concordance with the APOE genotyping.
Methods: One hundred seventy-eight specimens were analyzed using the Lumipulse® G ApoE4 and Pan-ApoE for the ApoE proteotype and evaluated its diagnostic concordance with the APOE genotype.
Results: The ApoE4 kit specifically detected the ApoE4 concentration in plasma samples, and the polymorphism could be classified clearly by the ratio of ApoE4 and Pan-ApoE amount in plasma.
Conclusions: The combination of Pan-ApoE and ApoE4-specific chemiluminescent enzyme immunoassay (CLEIA) assay is useful for predicting APOE ε4 allele status.
Apolipoprotein E (ApoE) is associated with lipid homeostasis by mediating the transport and clearance of lipids between organs via the plasma and interstitial fluids [1]. APOE mainly exists in 3 allelic variants, APOE ε2, ε3, and ε4, which differ in a single amino acid change at position 112 or 158; ApoE2 (Cys112, Cys158), ApoE3 (Cys112, Arg158), and ApoE4 (Arg112, Arg158) [1]. Among them, the APOE ε4 allele has been identified as the leading genetic risk factor for the sporadic form of Alzheimer’s disease (AD) [1–3]. Moreover, the presence of one or two copies of the APOE ε4 allele accelerates AD onset by approximately 10–20 years [2, 4]. Furthermore, the APOE ε4 haplotype is a major risk factor for amyloid-related imaging abnormalities (ARIA) in individuals treated with anti-amyloid therapies for AD [5, 6]. Indeed, the combined data from the EMERGE and ENGAGE clinical trials showed that 20.3% of APOE ε4 non-carriers and 43% of APOE ε4 carriers developed amyloid-related imaging abnormalities-edema (ARIA-E) in the high-dose group [5, 7]. In the Clarity AD clinical trial, 5.4% of APOE ε4 non-carriers and 15.8% of APOE ε4 carriers developed ARIA-E [6]. These data indicate that individuals who are APOE ε4 carriers are at greater risk of ARIA occurrence and severe ARIA-related adverse events [8, 9]. Therefore, there is a growing demand to identify the APOE ε4 haplotype.
In the study of APOE polymorphism, both genotyping and proteotyping methods have been used [10]. Among the numerous available genotyping techniques, including gene-based analyses such as polymerase chain reaction (PCR) plus sequencing, and real-time PCR [11, 12], HhaI restriction fragment length polymorphism (RFLP) analysis of a PCR-amplified domain of the APOE gene appears to be the conventionally adapted assay [13]. On the other hand, the characterization of the different ApoE proteotypes is carried out by isoelectric focusing and immunoblotting (IEF-IB), enzyme-linked immunosorbent assay (ELISA), mass spectrometric immunoassays (MSIAs), or biochip arrays [14–17]. Among them, IEF-IB is a commonly used assay method based on charge differences and separation of common isoforms [14, 18], but the main drawback of IEF-IB is that posttranslational modifications such as physiological sialylation/desialylation, diabetes-induced glycation or oxidative modification such as imine adduct formation can alter the charge, leading to the disagreement between genotype and proteotype [13, 14, 19]. In addition to the above, these genotyping and proteotyping assays pose several limitations; they can be time-consuming, technically complex, require additional sample processing, lack high-throughput capacity, or lack of fully automation. These technical disadvantages might be one of the reasons to hamper the implementation of APOE ε4 analysis in routine clinical practice.
In this study, we have developed fully automated chemiluminescent enzyme immunoassay (CLEIA) kit for ApoE4 and Pan-ApoE on the LUMIPULSE® platform using monoclonal antibodies recognizing ApoE proteins, including the ApoE4-specific antibody [15]. Here, we evaluated its diagnostic concordance with APOE genotyping and showed that the CLEIA ApoE4 and Pan-ApoE assays robustly distinguish between APOE ε4 carrier status in plasma.
The study included 178 Japanese volunteers, who were from our company and provided informed consent prior to enrollment. After the blood samples were collected into BD Vacutainer® K2EDTA tubes (Catalog number: 365900; BD Biosciences, NJ, USA), all tubes were gently inverted 8–10 times to avoid micro-clotting, and centrifuged at room temperature within 30 min (1,500× g for 10 min). The plasma layer was aliquoted into 2.0 mL polypropylene tubes (Catalog number: 72.694.600; Sarstedt, Numbrecht, Germany), followed by freezing in liquid nitrogen. Samples were kept at –80°C until use. Since the APOE gene is hereditary, collected samples were completely anonymized so that biographic and medical information about these participants was not collected. Study procedures were compliant with the principles of the Declaration of Helsinki. The ethics committee approved the study at H.U. Group Holdings, Inc. in the 2022 fiscal year (22-012-01).
ApoE4 and Pan-ApoE were sequentially analyzed using the Lumipulse® G ApoE4 and Pan-ApoE test (Fujirebio Europe N.V., Gent, Belgium). The LUMIPULSE® systems, e.g., LUMIPULSE® G1200 or G600II, are instruments using chemiluminescence quantitative sandwich immunoassay principle, with an assay time of 30 min per test. Samples were tested according to the instructions for use. The measurements were carried out on the LUMIPULSE® G1200 automated immunoassay analyzer with a single calibration run. Each assay required the following sample volume: 20 µL for ApoE4, and 20 µL for Pan-ApoE. For each analyte, one assay lot was used to measure all the samples.
The APOE genotyping with PCR-RFLP method was previously reported [20]. Briefly, the DNA was purified from peripheral whole blood, and the extracted DNA was used as the template for PCR. The PCR products were purified using QIAquick PCR Purification Kit (QIAGEN Inc., CA, USA) and the purified PCR products were digested with 10 units HhaI (New England BioLabs Inc., MA, USA) at 37°C for 2 h. For the restricted patterns, 20 µL of the digested fragments was separated by electrophoresis on a 15% polyacrylamide gel using SuperSepTMDNA (FUJIFILM Wako Pure Chemical Corporation, Japan) at 30 mA for 1 h. After electrophoresis, the gel was stained with ethidium bromide, and the polymorphic patterns were analyzed. Furthermore, the PCR fragments were sequenced by ABI PRISM® 310 Genetic Analyzer (PE Applied Biosystems, CT, USA) following the manufacturer’s instructions.
All statistical analyses were performed in version 4.2.2 of R (R Foundation for Statistical Computing, Vienna, Austria). Group comparisons for continuous variables were performed using the Wilcoxon rank-sum test with Bonferroni correction. Cross-table analyses were performed by χ2 test. The significance level of the statistical analyses was set at P < 0.05.
We conducted both PCR-RFLP and PCR plus sequencing, and confirmed that the genotype results from both methods are consistent. The distribution of APOE genotypes among the subjects in this study was summarized in Table 1; 78.7% (140/178) of subjects were APOE ε4 non-carriers (ε2/ε3 or ε3/ε3), and 21.3% (38/178) had at least one APOE ε4 allele (ε2/ε4, ε3/ε4, and ε4/ε4).
APOE genotype distributions in this study
| Genotype | n | Frequency |
|---|---|---|
| ε2/ε2 | 0 | 0.00% |
| ε2/ε3 | 24 | 13.48% |
| ε3/ε3 | 116 | 65.17% |
| ε2/ε4 | 9 | 5.06% |
| ε3/ε4 | 19 | 10.67% |
| ε4/ε4 | 10 | 5.62% |
| Total | 178 | - |
-: not applicable
All 178 samples were analyzed using the Lumipulse® G ApoE4 and Pan-ApoE. The concentration levels ranged from 0.0 µg/mL to 21.9 µg/mL for ApoE4 (Figure 1A), and from 7.5 µg/mL to 86.2 µg/mL for Pan-ApoE (Figure 1B). There was the trend that the ApoE4 concentration mostly dose-dependently increased with the copy number of APOE ε4 allele, allowing for the statistically significant (P < 0.001) distinction among these groups (Figure 1A). Although some samples in the APOE ε4 heterozygous group had higher ApoE4 concentrations which are comparable to the concentrations of the APOE ε4 homozygotes, the two groups were significantly different (P < 0.001) (Figure 1A). On the other hand, Pan-ApoE concentrations in APOE ε4 homozygous group was significantly lower than those in heterozygous and null group (P < 0.01 and P < 0.001, respectively) (Figure 1B), but there was no statistical difference in Pan-ApoE concentrations between heterozygous and null group. Furthermore, the ApoE4/Pan-ApoE ratio could clearly classify the APOE ε4 carrier status (Figure 1C and Table 2). The range of the ApoE4/Pan-ApoE ratio in this study was between 0.0% and 140% (Figure 1C). Then, we preliminary set the cutoff values at 5% and 75% in the ApoE4/Pan-ApoE ratio (Figure 1C), and cross-table analyses were performed by χ2 test, resulting in a significant difference (Table 2).
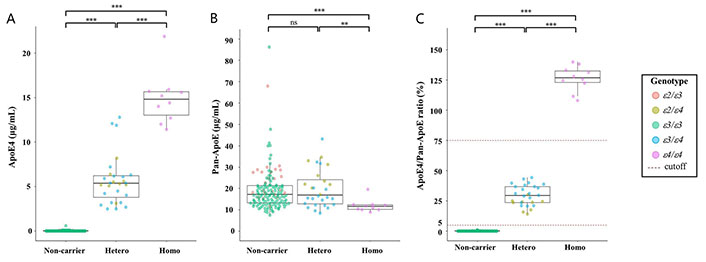
Distribution of biomarkers. Beeswarm boxplots of the assays ApoE4 (A), Pan-ApoE (B), and ApoE4/Pan-ApoE (C) by APOE ε4 carrier status. Boxplots display the median values with the interquartile range (lower and upper hinge) and ± 1.5-fold the interquartile range from the first and third quartile (lower and upper whiskers). Data were analyzed using Wilcoxon signed-rank tests with Bonferroni correction. ** P < 0.01; *** P < 0.001. ε2: APOE ε2; ε3: APOE ε3; ε4: APOE ε4; ns: not significant
Cross tabulation analysis of APOE ε4 genotype versus proteotype
| Category | -/- | ε4/- | ε4/ε4 | Total |
|---|---|---|---|---|
| -/- | 140 | 0 | 0 | 140 |
| ε4/- | 0 | 28 | 0 | 28 |
| ε4/ε4 | 0 | 0 | 10 | 10 |
| Total | 140 | 28 | 10 | 178 |
The horizontal cells are genotype, and the vertical cells are proteotype. χ2 = 356; df = 4; P < 0.0001
In our study, we established ApoE4 and Pan-ApoE assays on the LUMIPULSE® platform, and demonstrated their performances by setting cutoff values for the ApoE4/Pan-ApoE ratio to optimize its concordance with the APOE genotype. Consistent with previous reports [15], we found that the ratio could completely match the APOE ε4 allele status.
It has been reported that the levels of plasma ApoE decrease in a perfect parametric fashion across the six APOE genotypes (ε2/ε2 > ε2/ε3 > ε2/ε4 > ε3/ε3 > ε3/ε4 > ε4/ε4) [21]. In this regard, our results showed a similar trend (Figure 1B), although the APOE ε2 homozygotes were not obtained in this study because it is a rare variant [22]. On the other hand, the frequency of the APOE ε4 homozygotes or heterozygotes in this study was comparable to the previous reports [3, 23]. Further study with a lager sample size could address these points.
Both genotyping and proteotyping methods have been used for APOE polymorphism analysis [10]. However, it has been reported that the ApoE proteotype by the IEF-IB method can be susceptible to protein modification and degradation, resulting in discordant results of the proteotype-genotype correlation [13, 14, 19]. In this point, while the immunoassay examined in this study, ELISA [15], MSIAs [16], or biochip arrays [17] might show robustness toward both post-translational modification and protein degradation, the Lumipulse® G ApoE4 and Pan-ApoE assays could be better than the other proteotyping assays in terms of improved accuracy and reproducibility, elimination of technical skill influence, and reduced labor and costs.
Recently, it has been reported that the APOE ε4 carrier or non-carrier could be classified with automated immunoassay for ApoE4 [24, 25]. However, it might be difficult to separate homozygotes and heterozygotes completely by ApoE4 assay alone because peripheral blood ApoE levels might be affected by individual differences or even diets [26]. Indeed, some ApoE4 concentrations in the APOE ε4 heterozygous group were similar to those of the APOE ε4 homozygotes (Figure 1A). By contrast, the Lumipulse® G ApoE4/Pan-ApoE ratio could distinctly separate APOE ε4 status among null, heterozygote, and homozygote because Pan-ApoE could serve as an internal control to minimize potential variability of plasma ApoE4 concentrations. Furthermore, LUMIPULSE® G1200 can perform 120 tests/h, suggesting that this system is suitable for high-throughput screening. Additionally, the LUMIPULSE® platform has a product portfolio for AD-related biomarkers, allowing sequential measurement of several biomarkers [27–29].
The limitations of this study are the lack of various analyses using specimen information, the limited study population, and the lack of a validation cohort. Since the samples were collected from our internal volunteers, the genetic information related to AD needs to be protected from leakage, which affected the availability of the biomedical information about the study participants. In addition, the samples are collected from our internal volunteers aged between 20s and 50s, consequently suggesting no serious illnesses that might affect the modification of the ApoE protein. Therefore, further investigations using patient specimens with underlying diseases such as diabetes mellitus, would be necessary in future. Besides, the samples in this study were composed of Japanese volunteers, and it would be necessary to compare the results with other racial groups. Furthermore, the cutoff values which were used in this study, could not be validated in an independent secondary cohort, suggesting that further studies are required to address these points.
In conclusion, this study successfully demonstrated that the combination of ApoE4 and Pan-ApoE CLEIA assays was useful for predicting APOE ε4 allele status. It has been reported that APOE ε4 genotype is strongly associated with ARIA and exhibits its allele copy-number dependent effects [5–8]. Indeed, it is recommended to perform a test for APOE ε4 before initiating anti-amyloid treatments [7, 8]. Therefore, the Lumipulse® G ApoE4 and Pan-ApoE assays could contribute to risk discussions and safety considerations of disease-modifying therapies for AD.
AD: Alzheimer’s disease
ApoE: apolipoprotein E
ARIA: amyloid-related imaging abnormalities
CLEIA: chemiluminescent enzyme immunoassay
IEF-IB: isoelectric focusing and immunoblotting
PCR: polymerase chain reaction
RFLP: restriction fragment length polymorphism
We thank the participants of the studies who made this study possible. We are thankful to all the employees of Fujirebio HD group for their generous help, especially we thank Kazuyuki Umeda, Mario Denoyette, Jarne Delanote, and Maarten De Jonge for their efforts to develop the ApoE kits.
TY: Resources, Data curation, Investigation, Methodology, Writing—review & editing. RD: Resources, Data curation, Methodology, Writing—review & editing. DM: Resources, Methodology, Writing—review & editing. HS: Resources, Data curation, Investigation, Writing—review & editing. JK and TN: Resources, Investigation, Methodology. IV and KA: Supervision, Writing—review & editing. HN: Data curation, Formal analysis, Supervision, Investigation, Visualization, Writing—original draft, Project administration, Writing—review & editing.
TY, HS, JK, TN, and HN are employees of Fujirebio Inc. TY, RD, DM, and IV are employees of Fujirebio Europe N.V. KA is a board member of Fujirebio Inc.
Study procedures were compliant with the principles of the Declaration of Helsinki. The ethics committee approved the study at H.U. Group Holdings, Inc. in the 2022 fiscal year (22-012-01).
Informed consent to participate in the study was obtained from participants.
Not applicable.
Not applicable.
Not applicable.
© The Author(s) 2023.
Copyright: © The Author(s) 2023. This is an Open Access article licensed under a Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, sharing, adaptation, distribution and reproduction in any medium or format, for any purpose, even commercially, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
Janusz Wiesław Błaszczyk
Priyanka Sengupta ... Debashis Mukhopadhyay
Danqing Xiao, Chen Zhang
Carlos Gutierrez-Merino
Julius Mulumba ... Yong Yang
Felipe P. Perez ... Maher Rizkalla
